
Cell Cover | Shuimubio Aids in Deciphering Chloroplast Gene Transcription Machinery
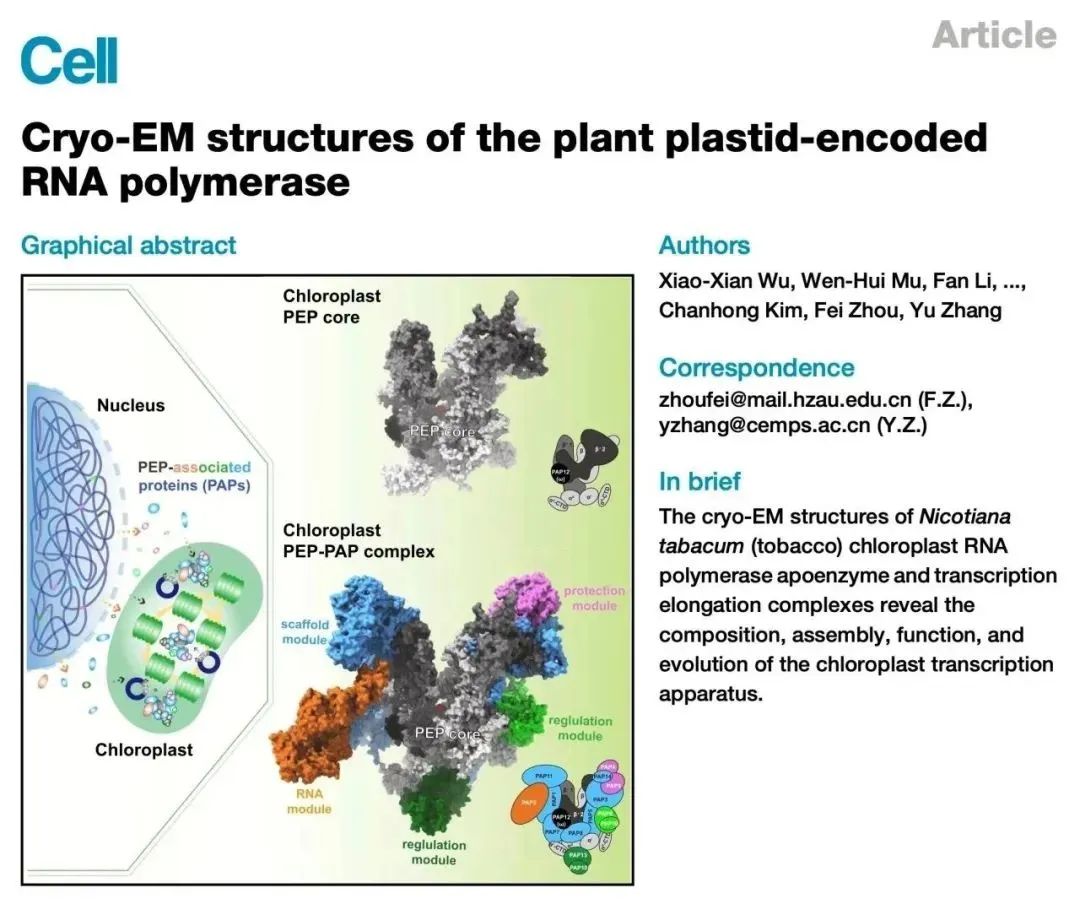
On March 1, 2024, the team led by Zhang Yu from the Molecular Plant Science Excellence Innovation Center of the Chinese Academy of Sciences, in collaboration with the team led by Zhou Fei from Huazhong Agricultural University, published a cover research paper titled "Cryo-EM structures of the plant plastid-encoded RNA polymerase" in Cell. Utilizing chloroplast transformation technology, the research team successfully established a method to purify endogenous PEP (plastid-encoded RNA polymerase) from tobacco leaves. They deciphered the high-resolution cryo-electron microscopy structures of the chloroplast RNA polymerase PEP complex and PEP transcription elongation complex, revealing the subunit composition, assembly mode, special functions, and evolutionary adaptation of the chloroplast gene transcription machinery. This research provides a structural basis for further studying the mechanisms and functions of transcriptional regulation in chloroplasts.
Background of the Research:
Photosynthesis in chloroplasts converts light energy into chemical energy, absorbs carbon dioxide, and releases oxygen, playing a crucial role in shaping the Earth's biosphere. Chloroplasts are believed to have evolved approximately 1.5 billion years ago through endosymbiosis with cyanobacteria. During evolution, chloroplast genes either became obsolete or gradually transferred to the cell nucleus, resulting in the retention of only 110-130 genes in the chloroplast genome of most land plants, most of which encode components involved in gene transcription, protein translation, and photosynthesis.
The chloroplast genome retains two types of RNA polymerases: the bacterial-type plastid-encoded RNA polymerase (PEP) and the phage-type nuclear-encoded RNA polymerase (NEP).
Chloroplast PEP plays a crucial role in the development of primitive plastids and serves as the primary RNA polymerase transcribing 80% of chloroplast genes in mature chloroplasts. The PEP core retains a structure similar to that of bacterial RNA polymerase, including 2 α subunits, 1 β subunit, 1 β'1 subunit, and 1 β'2 subunit, but lacks the ω subunit, with a total molecular weight of approximately 450 kDa. A unique feature of PEP is that several nuclear-encoded eukaryotic-origin PEP-associated proteins (PAPs) tightly interact with the PEP core and assemble into a supercomplex with a molecular weight of approximately 1 MDa. Each PAP mutation exhibits phenotypes similar to those of PEP core subunit mutants, displaying chloroplast developmental defects such as albino/pale phenotypes. Although extensive work has explored the composition and function of chloroplast PEP, unanswered questions remain regarding how eukaryotic-origin PAPs assemble with the bacterial-origin PEP core into complexes and how they regulate the transcriptional activity of PEP.
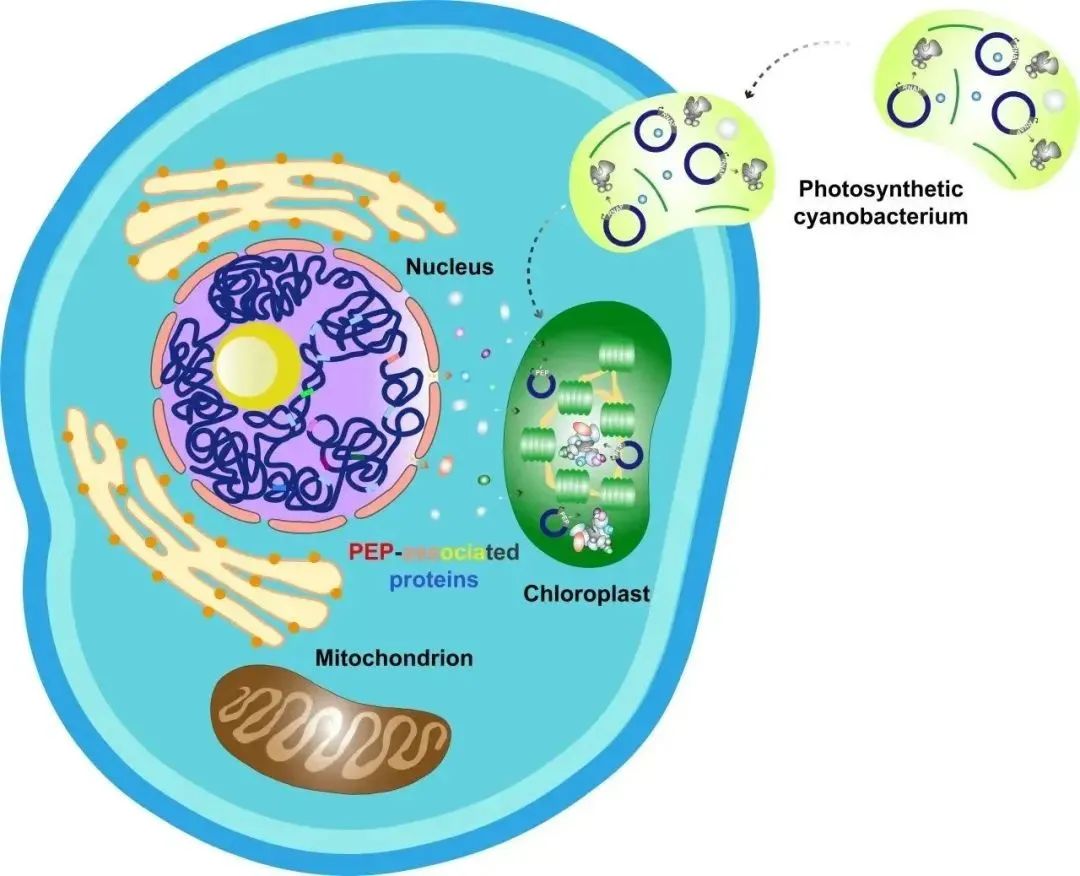 Figure 1: Evolution of Chloroplasts
Figure 1: Evolution of Chloroplasts
Research Content:
To purify endogenous tobacco PEP supercomplex, the research team constructed transgenic tobacco plants by inserting a DNA fragment encoding a tandem His-Flag tag at the 3' end of the chloroplast genome rpoC2 gene. Subsequently, purification steps such as affinity, ion exchange, and size-exclusion chromatography were performed using 30-45-day-old transgenic tobacco leaves. Finally, a highly purified PEP supercomplex with transcriptional catalytic activity was obtained.
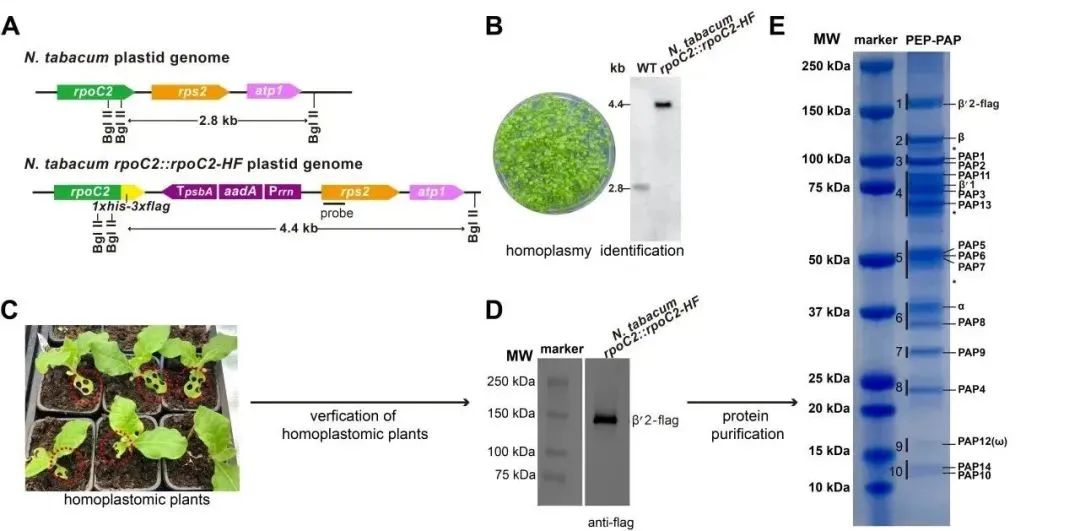 Figure 2: Purification Strategy of Tobacco PEP
Figure 2: Purification Strategy of Tobacco PEP
The structure of PEP reveals that the binding site of PAP12 is identical to the ω subunit of cyanobacterial RNA polymerase, with highly conserved primary sequence and three-dimensional structure, indicating that PEP contains a complete bacterial-origin RNA polymerase, constituting the "catalytic module" of PEP. Additionally, fifteen eukaryotic-origin PAPs bind to the periphery of the PEP core, forming different functional modules, including the "scaffold module," "protective module," "RNA module," and "regulatory module," making it the most complex gene transcription machinery known to date. The "scaffold module" consists of seven subunits (PAP1, PAP3, PAP5, PAP7, PAP8, PAP11, and PAP14), which interact extensively with PEP, occupying over half of the PEP core surface, and forming multiple inter-subunit structural domains with the PEP core, stabilizing the "catalytic module" and providing binding scaffolds for other modules. The "protective module" comprises a Fe-SOD heterodimer, composed of two subunits, PAP4 and PAP9 (also known as FSD3 and FSD2), which protect PEP from oxidative damage in chloroplasts through their superoxide dismutase activity. The "RNA module" consists of a single subunit, PAP2, which binds to the periphery of the RNA synthesis channel, specifically binding RNA in a sequence-specific manner, participating in and assisting in RNA post-transcriptional processing. The "regulatory module" comprises four subunits, PAP6, PAP13, and two PAP10s (also known as FLN1, FLN2, and Trx z), involved in protecting the C-terminal domain of the "catalytic module" α subunit and potentially regulating the transcriptional activity of PEP. These functional modules are all encoded by the nuclear genome, translated in the cytoplasm, and then transported to the chloroplasts to form complexes with the catalytic module of PEP, thereby controlling gene expression in chloroplasts by the nucleus.
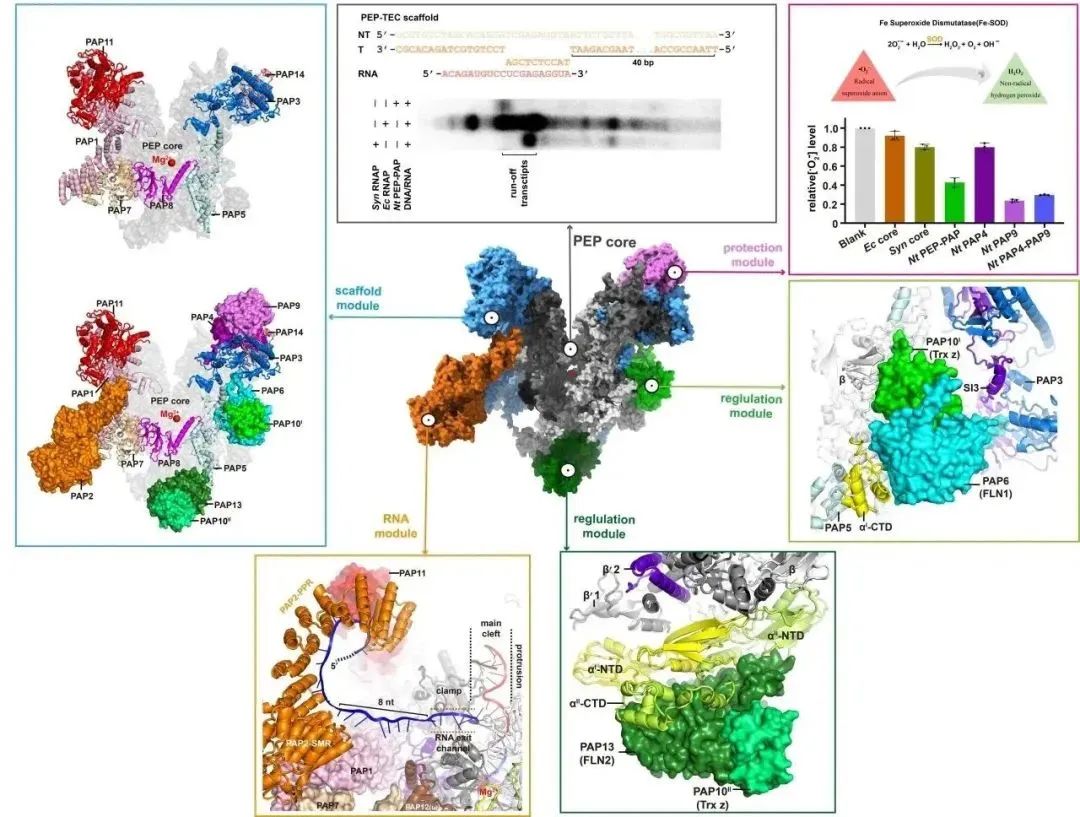 Figure 3: Composition and Function of Chloroplast PEP Modules
Figure 3: Composition and Function of Chloroplast PEP Modules
Finally, the research team conducted an evolutionary analysis of PEP-PAP protein subunits and found that the evolution time of the chloroplast gene transcription machinery PEP-PAP is basically consistent with the time of plant colonization of land. The chloroplast gene transcription machinery of land plants evolved unique functions and regulatory mechanisms by recruiting these additional subunits, helping them adapt to terrestrial environments and unique life cycles.
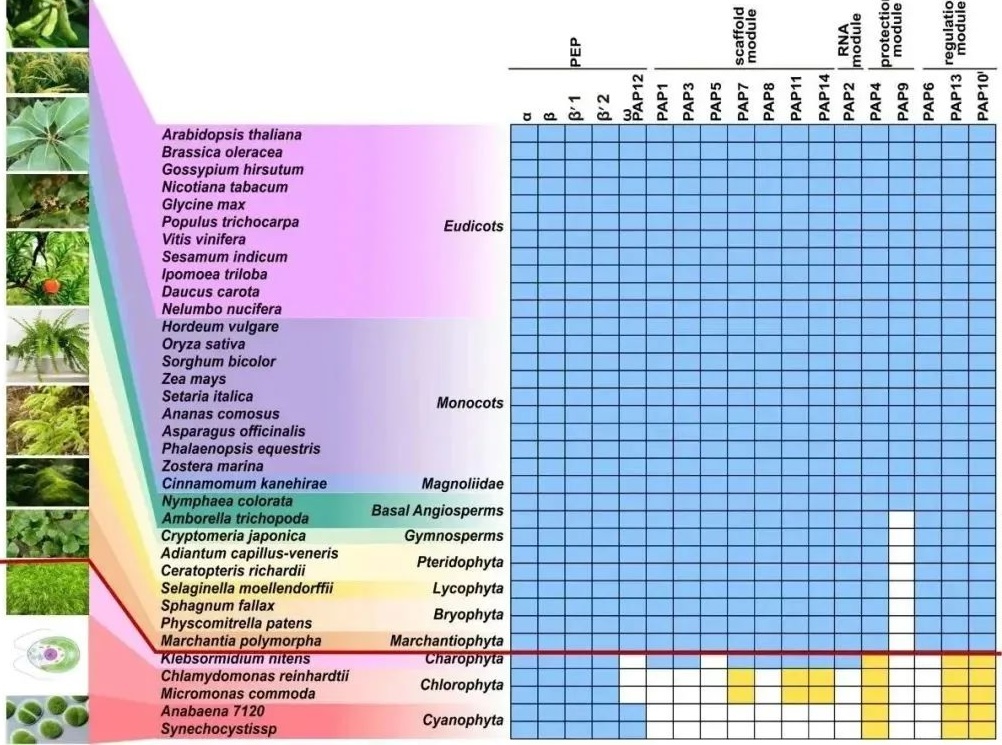 Figure 4: Evolution of Chloroplast PEP
Figure 4: Evolution of Chloroplast PEP
At the fundamental research level, this study lays the groundwork for further exploration of the working mode of the chloroplast gene transcription machinery, understanding the regulatory mechanisms of gene expression in chloroplasts, and remodeling the chloroplast gene expression regulatory network. At the application level of synthetic biology, this study provides a starting point for improving the efficiency of plant chloroplast bioreactors, thereby facilitating the production of recombinant vaccines, recombinant protein drugs, and natural products. Under the dual carbon goals of "peak carbon" and "carbon neutrality," this study provides new ideas for increasing the gene expression levels of photosynthetic systems, thereby aiding in the efficient carbon sequestration of plants.
Shuimubio CryoEM Platform
Shuimubio was deeply involved in the data collection process of this research, providing services with a 300kV cryo-electron microscope, and offering GraFuture™ RGO grids during the sample preparation stage.

DOI of the research: https://www.sciencedirect.com/science/article/abs/pii/S0092867424000631